How was your sleep last night? Were you able to go to bed early and clock in a solid 7 to 9 hours of sleep, or were you tied up with work and had to postpone your bedtime till the draft was written and the email sent?
On average, we human beings spend 8 hours per day sleeping, which amounts to a total of one-third of our lifetime. That is, for a person who spends 75 years on earth, 25 years is spent sleeping. With such a significant amount of time spent in bed, one couldn’t help but wonder, what would happen if we just shift the bedtime later by an hour and a half, while keeping the wake-up time fixed? Surely it will not have much of an impact?
Well, turns out it will.
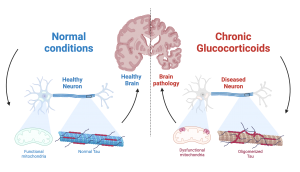
In a recent study published in Scientific Reports, Columbia postdoc Vikas Malik and colleagues found that for healthy females, after cutting sleep short by an hour and a half for a mere six weeks, cells that protect our blood vessels become negatively impacted. Evidence has shown that sleep restriction introduces a more pronounced risk for females than males, leading the researchers to focus solely on females for this study. With this reduction in sleep, the amount of harmful molecules (oxidants) starts to overpower that of the good molecules (antioxidants) in the cells. Since oxidants are harmful molecules which are responsible for producing the notorious “free radicals” and causing toxic effects in the body, and antioxidants good molecules responsible for detoxifying the body and repairing damages made by oxidants – this imbalance between the amount of oxidants and antioxidants will lead to impaired cell function and detrimental consequences in the form of cardiovascular diseases.
While the significance of sleep is well known, the mechanisms of how lack of sleep could impact human health has remained largely unclear. In fact, this study provides some of the first evidence demonstrating how mild chronic sleep deprivation could impact the health of our heart. In addition, contrary to previous studies that mainly examine sleep deprivation conditions in a compact time frame (e.g., acute sleep deprivation in flies for 10 days), the authors investigated how mild sleep restriction influences vascular cell health in females (i.e., pushing the bedtime 1.5 hours later than what the participants used to, while keeping the wake-up time fixed over the span of 6 weeks). Owing to a more “modern” work/life balance, the results from the authors’ experimental design carry higher relevance to our sleep patterns.
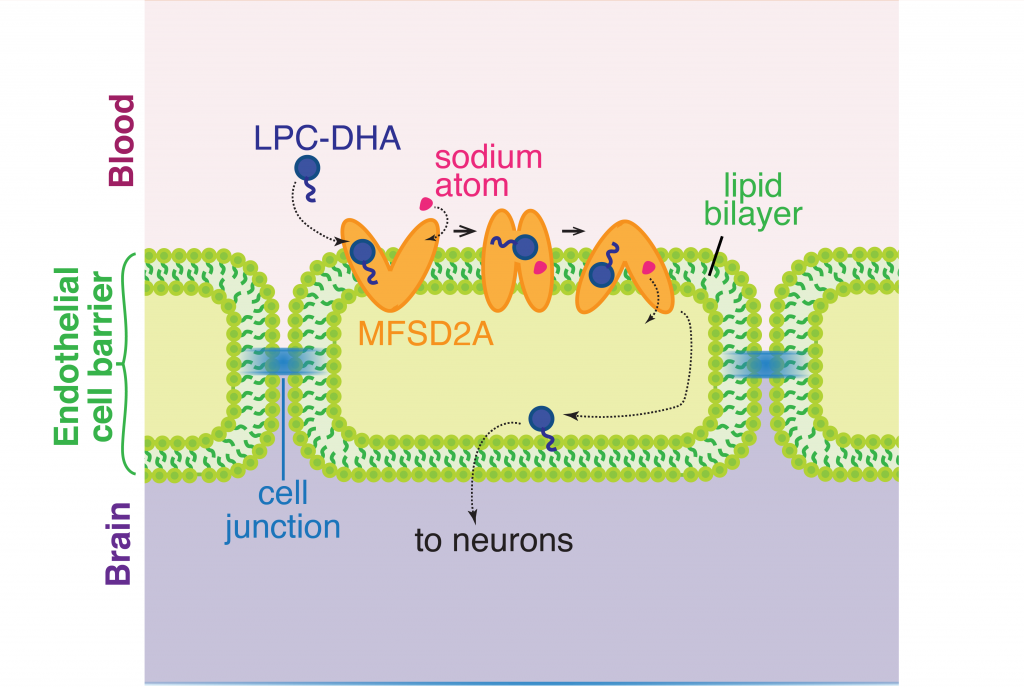
Specifically, the researchers found that after adequate sleep, the imbalance between oxidant (harmful) and antioxidant (good) molecules in the vascular cells can be cleared by a functional antioxidant response. This response is facilitated by a protein called serum response factor (SRF) – when sleep is adequate, the expression level of SRF increases to bring up the expression level of DCUN1D3, another protein which mediates antioxidant response of the cell. After sleep restriction, however, the antioxidant response cannot be turned on because the expression level of SRF is not adequate enough to activate a sequence of transcription of antioxidant genes, therefore hindering the restoration of balance between oxidant and antioxidant molecules in the cells (see illustration below).
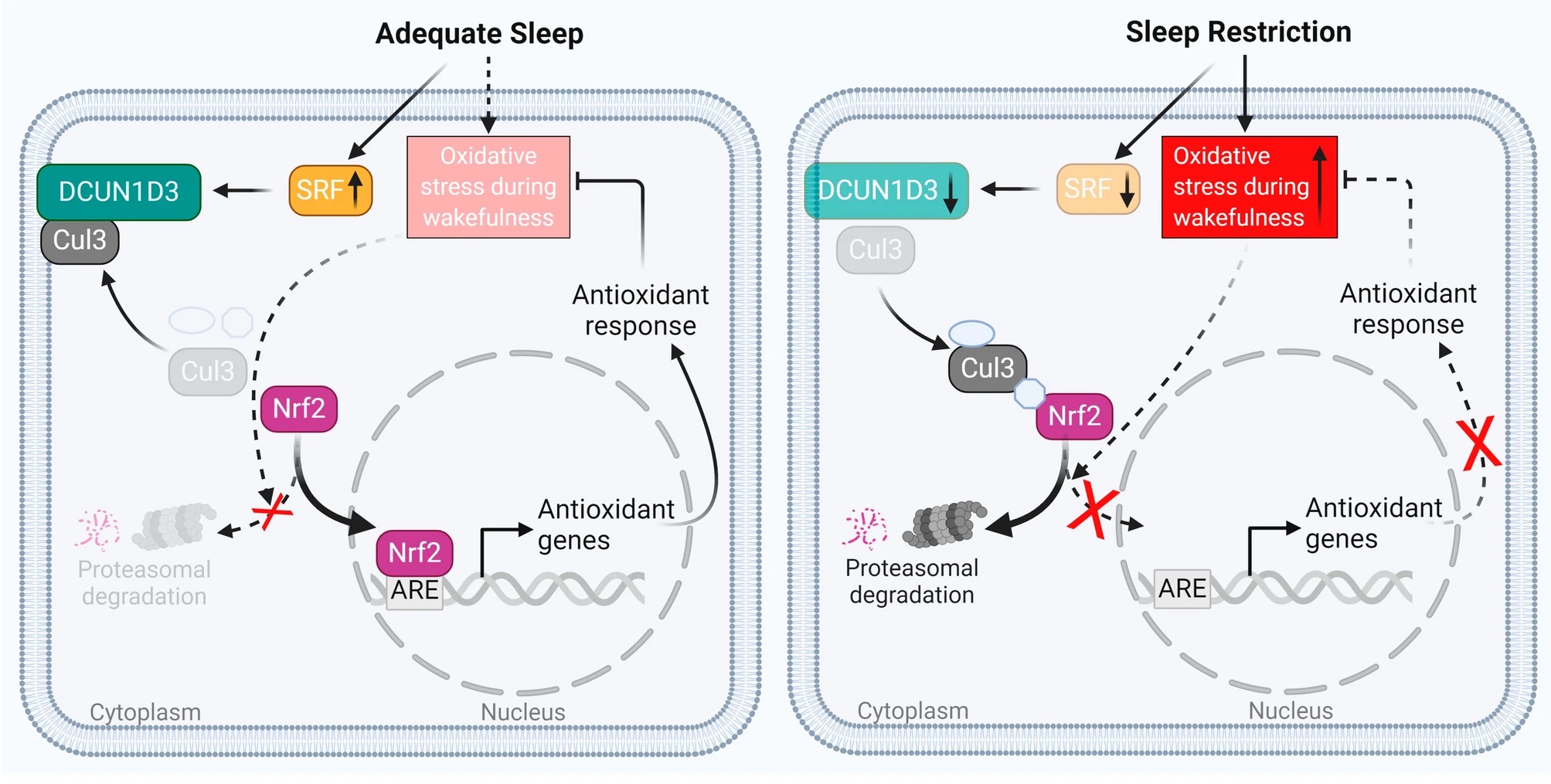
These findings carry significant implications on how we should schedule our sleep. While deadlines will always be looming and stress will always be present, maybe we should still make the effort to defend our bedtime. After all, as Shakespeare puts it, sleep is the “balm of hurt minds, great nature’s second course, chief nourisher in life’s feast”. Perhaps, unlike Macbeth, we should all strive to sleep more.
Written by: Linbi Hong
Reviewed by: Vikas Malik, Trang Nguyen, Martina Proietti Onori, Giulia Mezzadri, and Patricia Cooney