Camouflage is a common and essential survival strategy that is widely shared across the animal kingdom from reptiles, amphibians and fish, to birds and even some mammals. This ability allows them to hide in plain sight, but it is also a useful tool for hunting and a means of communication. Have you ever wondered how some animals can camouflage?

The dwarf cuttlefish (Fig.1) is renowned for its rapid and complex camouflage abilities, controlled by its brain. Five pairs of nerves emerge from the different brain regions and project to the different parts of the body (mantle, arms and fin) to regulate its skin’s chromatophores (these specialized cells contain different pigments, such as black, red, yellow, or brown, which contribute to the overall coloration of the skin).
Even though we usually crown the octopus as the master of disguise, the cuttlefish is, in fact, at the top of this game. The reason for this is that the cuttlefish’s skin possesses more color changing chromatophores per square inch than its fellow cephalopods (up to 200 per square millimeter to be precise). Recently, Columbia postdoc Tessa Montague and her colleagues introduced a groundbreaking brain atlas for the dwarf cuttlefish, *Sepia bandensis*. The team used a combination of Magnetic Resonance Imaging (MRI), deep learning, and histology to create this comprehensive neuroanatomical roadmap, which is now available online through the interactive web tool Cuttlebase (https://www.cuttlebase.org/). This study aims to understand how the cuttlefish brain processes visual information to achieve camouflage. Interestingly, despite that the brain has a global volume close to the one of a mouse (94% similarity), its organization is widely different. For example, the optic lobes that only represent a few percent in the mouse brain comprise 75% of the brain in the cuttlefish, indicating their crucial role in visual processing and behavior related to camouflage and environmental interaction. The research revealed significant insights into the neural basis of cuttlefish behavior and offered a valuable resource for further studies on neural representation and brain function.
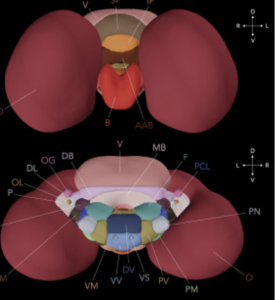
To achieve this, the team scanned the brains of eight cuttlefish using MRI. In combining manual segmentation with deep learning techniques to extract each brain from its surrounding tissue, they created the first merged, annotated template brain (Figure 2).
With such 3D model of brains like Cuttlebase, scientists can target brain regions to investigate precisely the function in the camouflage behavior.
The 3D and histological brain atlases presented here are invaluable tools for studying how cephalopods, like the dwarf cuttlefish, control their behavior through their brains.
With the help of the interactive platform Cuttlebase, researchers can precisely guide instruments for brain experiments and identify specific brain regions for their studies. These atlases also allow scientists to explore how genes and neural activity are distributed across different areas of the brain.
The brain atlas not only helps in understanding cephalopods but also serves as a resource for comparing the brain structures of different species. While little is known about the biology of the dwarf cuttlefish (Sepia bandensis), the common cuttlefish (Sepia officinalis) has been extensively studied to understand behavior, learning, memory, and skin pattern control. Despite the differences in their sizes, behaviors, and habitats, the data shows that their brain structures are quite similar. For example, Sepia bandensis lives in the Indo-Pacific coral reefs and uses high-frequency skin patterns to blend into its tropical environment. On the other hand, Sepia officinalis lives in the rocky waters of Europe and uses larger patterns to resemble rocks. Both species display unique aggression patterns during social interactions: Sepia officinalis shows a zebra pattern, while Sepia bandensis has a stippled pattern. These differences in behavior might be better understood through studies of neural activity.
This atlas is also useful for understanding evolutionary processes by comparing species that are evolutionary distant but still share similar anatomical organization. Thus, a recent study shows that despite about 600 million years of evolution, cuttlefish brains share some features with those of fruit flies. For instance, both have a central brain with two large optic lobes on the sides and learning and memory centers. Recent studies even suggest molecular similarities between certain brain cells in octopuses and fruit flies, highlighting the evolutionary connections.
This comprehensive brain atlas, combined with molecular and cellular maps, offers a deeper understanding of how these intelligent invertebrates evolved their complex cognitive abilities and behaviors. Comparing this atlas to those of other species, like the nematode C. elegans or the Human Connectome Project, provides valuable insights into brain function across different phyla. Such comparisons are crucial for uncovering universal principles of neural organization and function as well as the distinct evolutionary paths and ecological requirements of mammals and cephalopods reflected by neuroanatomical differences.
The creation of detailed brain atlases is foundational for advancing our knowledge of brain anatomy and behavior across the animal kingdom. These studies highlight the critical role of brain atlases in exploring neural structures and functions, significantly contributing to neuroscience. Despite the existing atlases, there is a pressing need for more to fully understand brain evolution throughout the animal kingdom.
By expanding the collection of brain atlases across a wider range of species, researchers can uncover evolutionary pathways and adaptations that have shaped cognitive abilities. The detailed information provided by these atlases helps correlate brain structure with function and behavior, offering insights into the neural basis of complex traits. Increasing the number of brain atlases will enable scientists to trace the development of neural mechanisms from simple organisms to complex beings, enhancing our understanding of brain evolution and capabilities.
Investing in the creation of more brain atlases will build a robust framework for comparative neuroanatomy, deepening our understanding of individual species and illuminating broader evolutionary trends. The insights from these comparative studies are crucial for developing new hypotheses about brain function, improving neural models, and advancing our knowledge in neuroscience and related fields. If you want to know more details check out the original paper.
Written by: Nathalie Houssin
Reviewed by: Trang Nguyen, Margarita Angelova, Jerry Gourdin

 Figure 1: The number of infections in the households is closely followed by the number of hospitalizations reported. The blue line is a measure of the infection rate of a household and the gray region around it is the variation in that rate.
Figure 1: The number of infections in the households is closely followed by the number of hospitalizations reported. The blue line is a measure of the infection rate of a household and the gray region around it is the variation in that rate. Figure 2: All possibilities of transmission among the three age groups were tested in the full within household transmission model.
Figure 2: All possibilities of transmission among the three age groups were tested in the full within household transmission model.