“Eat what you want and stop when you’re full.”
For some people, this statement is absolutely invalid as they never feel full; they don’t have an ‘off-switch’ while eating. Sometimes, consuming food makes them feel even hungrier. These are some classic symptoms of binge eating. Binge eating falls under the big umbrella of eating disorders, which are serious mental health conditions characterized by persistent alteration of eating behavior and associated emotions. Three different diseases belong to the spectrum of eating disorders: anorexia nervosa, bulimia nervosa, and binge eating disorders. Although binge eating disorder is the most prevalent, surprisingly it does not get as much media coverage compared to anorexia and bulimia.
Binge eating results from “hedonic hunger” the drive to consume food not because of an energy deficit, but for the inherent pleasure associated with eating. The pleasure signal for bingeing relies mostly on the reward-associated component of feeding and sensory stimuli such as smell and taste. The reward system functions by raising the level of the neurotransmitter dopamine in a midbrain structure called the ventral tegmental area. Years of research in laboratory animals also depicted a positive correlation between binge eating and increased dopamine release. The endocannabinoid system has been connected with this rewarding aspect of food intake and represents the key system modulating bingeing. Fun fact, cannabis consumption leads to overeating (read: munching) by tricking the brain into feeling like it’s starving when in reality that’s not the case. In association with the endocannabinoids and reward system, the gut or gastric lumen also plays as a master driver controlling feeding behavior in general along with binge eating. Intriguingly, endocannabinoids are functionally dependent on the vagus nerve innervating the gastrointestinal tract. Overall, scientists have just begun to understand the complex nature of binge eating from the neurobiological and psychological standpoint.
A recent preprint by Dr. Chloé Berland and colleagues dissected, for the first time, the indelible role of the reward system, gut-brain axis, and endocannabinoids in binge eating. This study successfully leveraged a unique binge eating model in which a highly palatable milkshake was provided to mice in a time-locked manner. This binge-eating model was driven by reward values rather than metabolic demand, as animals had unlimited access to less palatable food throughout the test, so milkshake consumption occurred in absence of energy depreciation. This study pinpointed that two phases of binge eating, anticipatory and consummatory, are controlled by a specific dopamine receptor called D1 (D1R).
The cannabinoid receptors are available both in the peripheral and central nervous systems. The current study aimed to uncover the specific connection between peripheral cannabinoid and bingeing. To achieve that goal, a peripherally restricted chemical was administered in the mice to block the activity of the cannabinoid receptor. Dr. Berland and her colleagues observed that the injection of peripheral cannabinoid blocker completely silenced the hedonic drive for bingeing. This finding reveals that physiologically, the peripheral endogenous cannabinoid acts as a gatekeeper for binge eating.

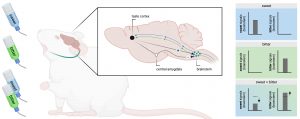
Figure 1. Schematic representation showing how the peripheral endocannabinoid mediates bingeing via the gut-brain axis. The left panel of the diagram shows that increased peripheral endocannabinoid causes increased reward and bingeing while the right side shows the opposite. The brain region, Nucleus Tractus Solitarius (NTS), Endocannabinoids, or eCB (2-Arachidonoylglycerol molecule as a representation). Adapted from Berland et al. and created with Biorender.com.
To delve more into the involvement of the gut-brain axis in endocannabinoid-mediated bingeing, the current study used vagotomy, a severing of the vagus nerve’s connections to the gastrointestinal tract and other abdominal organs, to shut off the function of the vagus nerve in these organs. Injection of peripheral cannabinoid blocker in vagotomized mice led to strong activation of a brain region known to play a key role in receiving signals from the gut about meals, the nucleus tractus solitarius (NTS) (see Figure 1). This observation indicates that the peripheral endocannabinoids are important influencers that act in between the gut and brain in regulating the hedonic drive for food.
This study took advantage of a unique, cutting-edge technology called fiber photometry to further dissect how the endocannabinoids control the reward component of bingeing. With fiber photometry, the neural activity of specific brain regions can be detected in awake animals. The neural activity in the midbrain reward area was dampened after the peripheral endocannabinoid blocker injection. This finding suggests that peripheral endocannabinoids control the food craving by modulating the reward system.
Taken together, the observations of this study provide crucial mechanistic insights on gut-brain and endocannabinoid integration. Using state-of-the-art tools, this study sheds light on the previously unexplored regulatory mechanism of the endocannabinoids in bingeing. So, the next time you binge eat a pint of Ben & Jerry’s ice cream, you know it’s not only the burst of pleasure chemical dopamine but also your body endocannabinoids tricking your gut and brain to finish it all.
These new and exciting data warrant that peripheral endocannabinoid blockers could be utilized for the treatment of binge eating disorders or related eating disorders in humans. Patients with eating disorders struggle mentally, emotionally, and physically. For instance, individuals with eating disorders often become victims of body shaming. We can always do more to help binge eating disorder patients in the recovery process. Here are some useful resources for patients struggling with binge eating disorder:
https://www.nationaleatingdisorders.org/
Dr. Chloé Berland is a Postdoctoral Research Scientist in the Department of Preventive Medicine where she studies the effect of overfeeding on brain circuits. She also serves as CUPS secretary.