Cell death is an important process through which the structure of our bodies are shaped throughout development. For example, soft tissue cells between the fingers and toes undergo apoptosis (programmed cell death) to separate the digits from each other during development (Figure 1). Billions of cells die in our bodies every day and a prompt clearance of dead cells and their debris is important for maintaining tissue homeostasis. Tissue homeostasis requires a very tight control of the balance between cellular proliferation and differentiation. The majority of these dead cells are cleared by macrophages, a type of immune cell, through a process called “efferocytosis”.
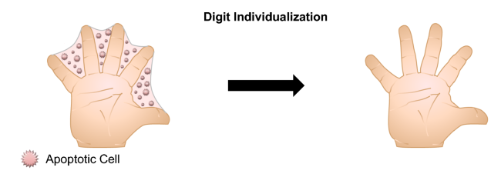
Figure 1: Programmed cell death is an important process during development that serves to remove superfluous cells and tissues. Figure was adopted from “Mechanical Regulation of Apoptosis in the Cardiovascular System”.
If dead cells are not appropriately cleared by macrophages, they start leaking material in the cellular environment that causes inflammation and tissue damage. Efficient efferocytosis prevents this from happening, and thereby protects tissues from inflammation. Macrophage-mediated efferocytosis is an important process to promote the resolution of inflammation and restore tissue homeostasis. While inflammation causes swelling, redness, and pain, efferocytosis does not. In fact, enhancing efferocytosis has the potential to dampen inflammation and reduce tissue necrosis which is caused by injury or failure of the blood supply. Defective efferocytosis contributes to a variety of chronic inflammatory diseases such as atherosclerotic cardiovascular disease, chronic lung diseases, and neurodegenerative diseases. Understanding the mechanisms that regulate efferocytosis could help us develop novel therapeutic strategies for diseases driven by defective efferocytosis and impaired inflammation resolution.
Like other cells in the body, macrophages need energy to maintain their activity. Glycolysis and oxidative phosphorylation are two major metabolic pathways to provide energy for cells. Glycolysis is a process in which glucose (sugar) is broken down through enzymatic reactions to produce energy. Macrophages take up glucose via glucose transporters on the cell surface, such as GLUT1. Glucose will be broken down to generate ATP (energy) and lactate, an end product of the glycolysis pathway.
A research group in the department of Medicine at Columbia University led by Dr. Maaike Schilperoort, a postdoctoral research scientist in Dr. Ira Tabas’ laboratory, identified a novel pathway in which efferocytosis promotes a transient increase in macrophage glycolysis via rapid activation of the enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2), a key enzyme in the glycolysis pathway to convert glucose to lactate (Figure 2).
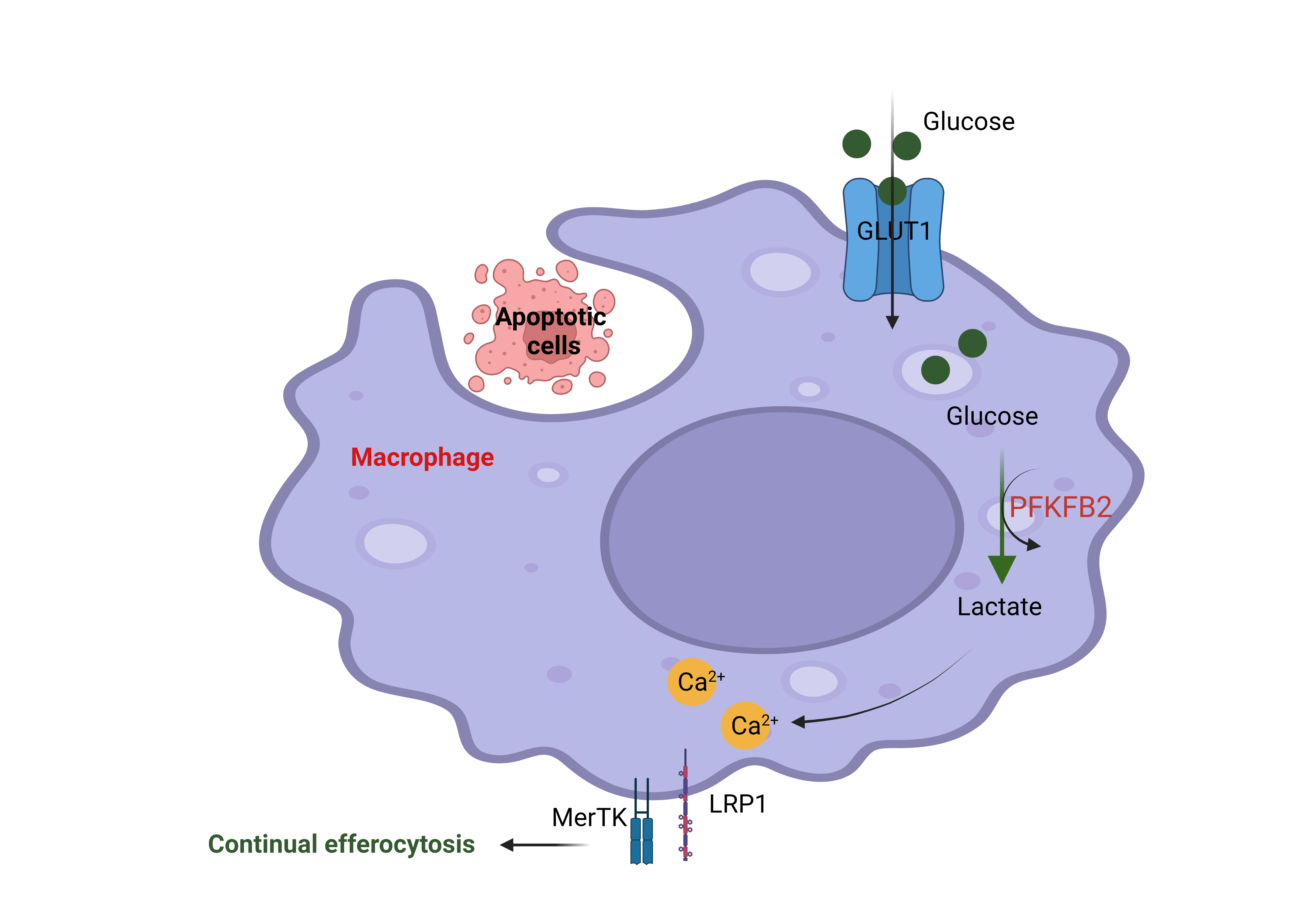
Figure 2: The engulfment of apoptotic cells by macrophages through efferocytosis increases glucose uptake via the membrane transporter GLUT1. Glucose is broken down into lactate through glycolysis, and this process is boosted by efferocytosis through activation of the enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2). Lactate subsequently increases cell surface expression of the efferocytosis receptors MerTK and LRP1. These efferocytosis receptors facilitate the subsequent uptake and degradation of other apoptotic cells in the tissue. This figure was created using Biorender.
MerTK and LRP1 are so-called “efferocytosis receptors” that allow the macrophages to bind to dead cells before they can engulf and degrade them. The current study found that the production of lactate leads to an increase in MerTK and LRP1 on the cell surface in a calcium-dependent manner to drive continual removal of dead cells (Figure 2). The authors mentioned that lactate promotes an efferocytosis-induced calcium-raising mechanism that could be involved in the mitochondria division. The mechanism of how lactate promotes the increasing of calcium is not well understood and needs to be explored more in the future. This finding provides potentially new therapeutic strategies for improving cell death clearance such as targeting an endogenous inhibitor of PFKFP2. This novel finding was published in Nature Metabolism in February 2023.
Reviewed by: Maaike Schilperoort , Erin Cullen, and Sam Rossano