Brains come in a variety of sizes. Several orders of magnitude separate the convoluted human brain from that of the fruit fly. But no matter the configuration, neuroscientists strive to study neurons in further and further detail, ideally at the single-cell level. However, consistently identifying individual neurons is not an easy endeavor; in most organisms there are so many neurons that even when labelled with colored tags we can’t see the forest for the trees.
Enter Caenorhabditis elegans, a worm with only 302 neurons. This tiny transparent worm comes in handy for scientists because they can easily follow the fate of any of its cells from the embryonic stage to adulthood. However, despite the somewhat predetermined developmental paths, the exact spatial location of each cell is slightly variable. Therefore, studies examining individual cell identities are doomed if they rely exclusively on relative position within each group of neurons.
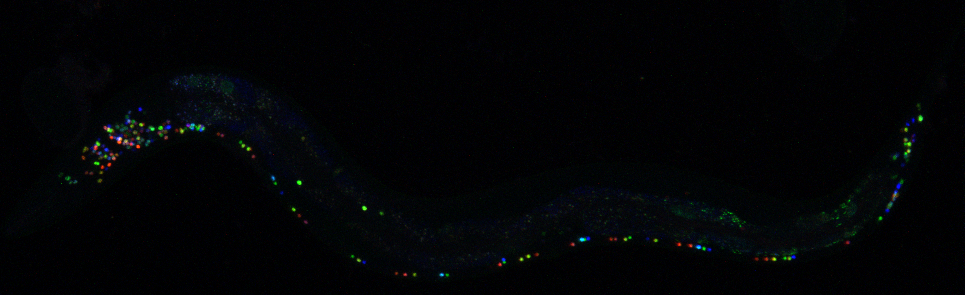
In their recent paper, Dr. Eviatar Yemini and colleagues introduce a solution to this problem in the form of deterministic fluorescent labelling of all 302 neurons of the worm C. elegans. Their approach, NeuroPAL (a neuronal polychromatic atlas of landmarks), produces the same pattern of colors across worms, which makes a fundamental improvement to previous similar techniques like Brainbow that produce random patterns.
The first challenge they faced was determining how many distinguishable colors they needed to correctly identify all neurons. Fortunately, the neurons of this worm are dispersed over its whole body, grouped in 11 different clusters, so they didn’t need all 302 to be labelled with distinct colors. The biggest of these ganglia contains roughly 30 neurons, so aiming for a range of colors around that number was enough. How do you get those colors? One could achieve a sizable palette by using just a few different fluorescent markers or “fluorophores” that are detectable at different intensities. However, selecting the final fluorophores wouldn’t have happened without a great example of scientific collaboration. Dr. Yemini had been struggling with the colors blending together until he contacted a colleague who told him about a new fluorophore, and this ended up being the missing puzzle piece he needed to achieve all the distinguishable fluorophores. Once they had carefully selected the three distinct markers, a clever trick of imaging them in red, blue and green, allowed them to obtain a whole RGB palette of colors.
The next step was to achieve the different levels of each fluorophore for each neuron in a consistent way across worms by changing the signal driving the fluorophores’ expression. Starting from a list of 133 previously published genes with different patterns of neuronal expression, Dr. Yemini tried them all, painstakingly narrowing down the list to 41 winners by a process of iterative trial and error, checking the resulting color combinations and whether the neurons could be distinguished at each step. This process alone spanned more than two years, and required deep dives into the literature and some expert judgement calls: “in one rather desperate case, I guessed the expression from the behavioral phenotype and, very luckily, was approximately right” Dr. Yemini says.
Once they achieved the final combination of colors, they had finally created a genetically modified worm that could pass on this colorful and robust pattern for many generations.
NeuroPAL is not only a technical feat in and of itself, but was also a creative outlet for Dr. Yemini, who enjoyed the highly collaborative art-meets-science project:
“In high school, I had to choose between applying to art school or following what I thought was a more traditional route. I loved the artistic process, I’d taken many art classes and even managed to score a scholarship for an after-school art program at SUNY Purchase. But I let luck guide my fate and ended up taking the non-artistic route. I really miss that part of me. The process of making NeuroPAL has felt like a taste of a part of me that I’d lost.”
So what do you do after you create a tool that allows you to unambiguously identify all neurons? You use it to explore more questions! Dr. Yemini and his colleagues applied their shiny new worms to study many old questions in the field. For example, they leveraged the individual cell identification to refine whole-brain activity imaging, with which scientists previously had the issue of being unable to determine neuronal identity. They succeeded in recording responses to different chemical stimuli, both attractive and repulsive, confirming previous results and adding new neurons to the response pathways, thus unraveling more complex neuronal networks. On the whole, they show that this new tool can be used for exploring a variety of questions in C. elegans.

You might be thinking, “This is all very cool, but what does a tiny transparent worm have to do with me?” While studying C. elegans in itself can shine light on some basic biological processes, it can also open the door to discoveries in more complex organisms and those more similar in neural organization to humans. Indeed, the authors suggest that this approach to unequivocally label cells could be translated to other models that are widely used in biomedical research, such as the fruit fly, fish and even mice. We still have a long way to go before we can create an entire rodent with a consistent pattern of shiny cells, but local labelling may be a more attainable goal. And eventually, this research will help us distinguish the trees from the forest.
Dr. Eviatar Yemini is an Adjunct Associate Research Scientist in the Department of Biological Sciences at Columbia University (Hobert lab). He will be starting his own lab at the University of Massachusetts Medical School in January 2022. Reach out to him for exciting job opportunities!