The brain and eyes develop through constant circulation of nutrients through the blood-brain and blood-retina barriers. One such nutrient that is essential for development is an omega-3 fatty acid called docosahexaenoic acid (DHA). DHA makes up a fifth of all the fatty acids required on the membranes of cells in the central nervous system. Neither the neurons in the brain nor the cells in the eye are capable of synthesizing DHA by themselves and therefore depend on dietary sources for DNA. Previously, scientists knew from cellular clues that this fatty acid most likely passed through to the blood-brain and blood-retina barriers in the form of lysophophatidylcholine (LPC-DHA) using a molecular channel. This transporter is known as a major facilitator superfamily domain containing 2A, or MFSD2A, with the help of sodium atoms regulating the channel. However, it was not clear how this channel allowed the passing of complex molecules like DHA. A recent study by Dr. Rosemary Cater and colleagues at Columbia University provided precise clues to further show the structure of this channel.
To investigate the structure of MFSD2A, the authors used a state of the art imaging technique called single-particle cryo-electron microscopy. This is a method of electron microscopy where a beam of electrons is transmitted through a rapidly frozen purified molecule. Because the sample is flash frozen, the molecules trapped in a frozen state can be imaged in their native shape as present in the cell and from multiple angles. By capturing and combining multiple captured 2D images, a 3D structure of the protein can be reconstructed with extreme accuracy. Cryo-electron microscopy is so impactful in biological significance that this method was awarded the 2017 Nobel Prize in Chemistry. found a number of molecular patterns and arrangements of protein chains that make up a full molecule of MFSD2A
Protein structure studies are typically among the most challenging grounds to explore in biology because proteins need to be captured in their native state as present in the cell. Past discoveries of various protein structures have been so instrumental in shaping therapeutic areas that the extent of mechanistic understanding of biological molecules has resulted in recognition by Nobel committees. Most recently, the discovery of the structure of the ribosome opened up fields of exploration into therapeutic interventions into ribosome diseases, some of which can lead to cancer.
To get the best chance at imaging the structure of MFSD2A, the scientists extracted and examined purified versions of this protein obtained from multiple organisms: the zebrafish, the frog, European cattle, the domestic dog, the red junglefowl, mice and humans. Finally, the authors found that the protein obtained from the red junglefowl, which is a rooster species that originates from Southeast Asia, was the most experimentally stable and most alike (73% identity) the human version of MFSD2A.
Using additional accessory proteins to help with the orientation of MFSD2A, the authors obtained high-quality images, with a resolution of 0.3 nanometer, or 0.3 billionth of a meter. From the imaging data, the authors found that MFSD2A protein itself is about 5 nm wide and 8 nm long. MFSD2A is a transporter protein and like many transporters, it contains repeated bundles of helices made of protein chains that traverse the cell membrane and are connected by a protein chain that loops within the space in the cell.
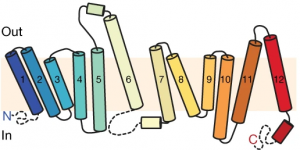
The cell membrane consists of two layers of lipid molecules, known as the lipid bilayer, that allow entry and exit of materials from the cell. These loops provide the shape to the protein inside the cell such that it appears to provide a large enough cavity opening from the lipid bilayer into the cellular space to allow the target molecules to enter the cell. Amino acids are the building blocks of proteins and the cavity contains amino acids of both water-attracting and water-repelling kind. This property makes it possible for many molecules of differing chemical nature to be able to be accommodated within the cavity. This cavity contains three important regions that allow for the protein to be specific and functional: a charged region, a binding site for sodium atoms and a lipid-specific pocket. The authors speculate that these parts help in establishing the mechanism by which LPC-DHA is transported from the outside into the cell. The multiple protein helices form two protein domains that capture LPC-DHA from outside the cell layer of the blood brain barrier of endothelial cells, then rock over a rotation axis so that now their confirmation switches and finally, they release the protein molecule into the cell. For this activity of movement of LPC-DHA, sodium atoms are absolutely required to allow for the shape change of the protein. Once LPC-DHA enters the barrier cells in this manner, the protein is then transported across to the other side of the cell facing the brain containing neurons.
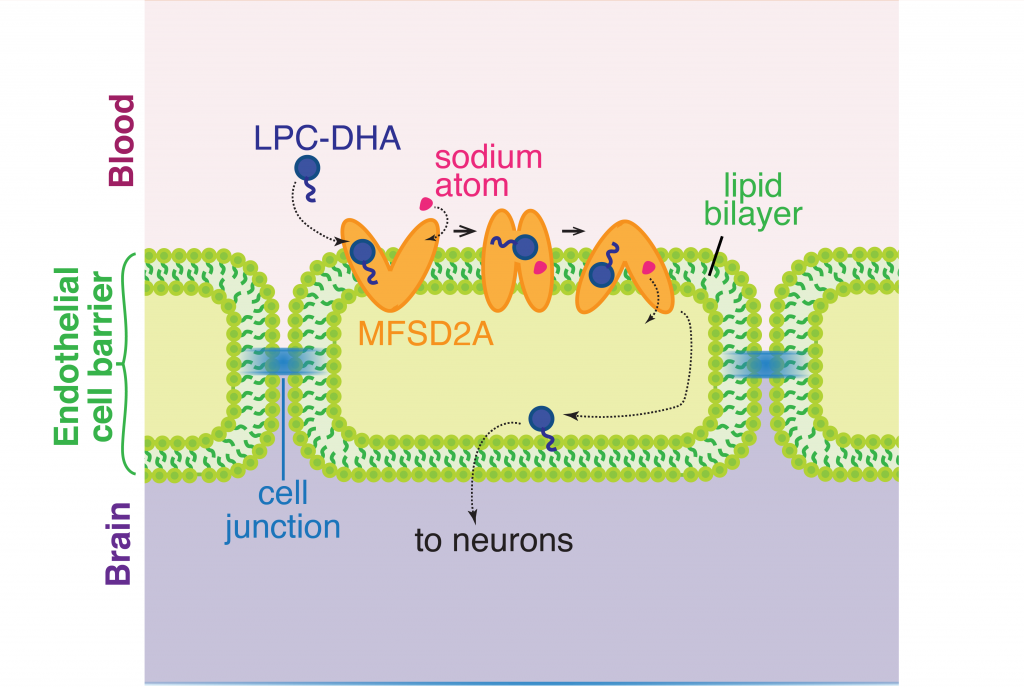
Humans with mutations in MFSD2A gene have abnormal brain defects such as microcephaly, and disruption of the gene in mice affected neuronal branching and fatty acid composition in the brain. The discovery of the structure of a molecule that mediates uptake of essential nutrients across the blood-brain and eye-brain barriers will help in the delivery of therapies of neurological diseases.
Dr. Rosemary J. Cater is a postdoctoral researcher in the lab of Dr. Filippo Mancia in the Department of Physiology and Cellular Biophysics at Columbia University.