Transposons, also known as mobile genetic elements, are segments of DNA capable of moving within the genome. This mobility can potentially induce mutations, as transposon “jumping” within a gene may interrupt the gene and trigger its loss. Transposons utilize their own enzymes, transposases, to move in and out of the genome.
Due to the potential harm caused by transposons, cells continually evolve strategies to control them. In response, transposons adapt, arming themselves with new strategies to evade cellular control. Mobilized transposons can hijack adjacent DNA information and transfer it to a new location, sometimes involving a gene that provides a survival advantage, thus increasing the transposon’s chances of propagation. Simultaneously, cells “steal” genes from transposons, repurposing them to function in processes advantageous to the cell. This ongoing arms race between transposons and host defense systems has facilitated a widespread exchange of genes between different life forms and is a major driver of genome evolution across all domains of life. Transposons play a crucial role in this process.
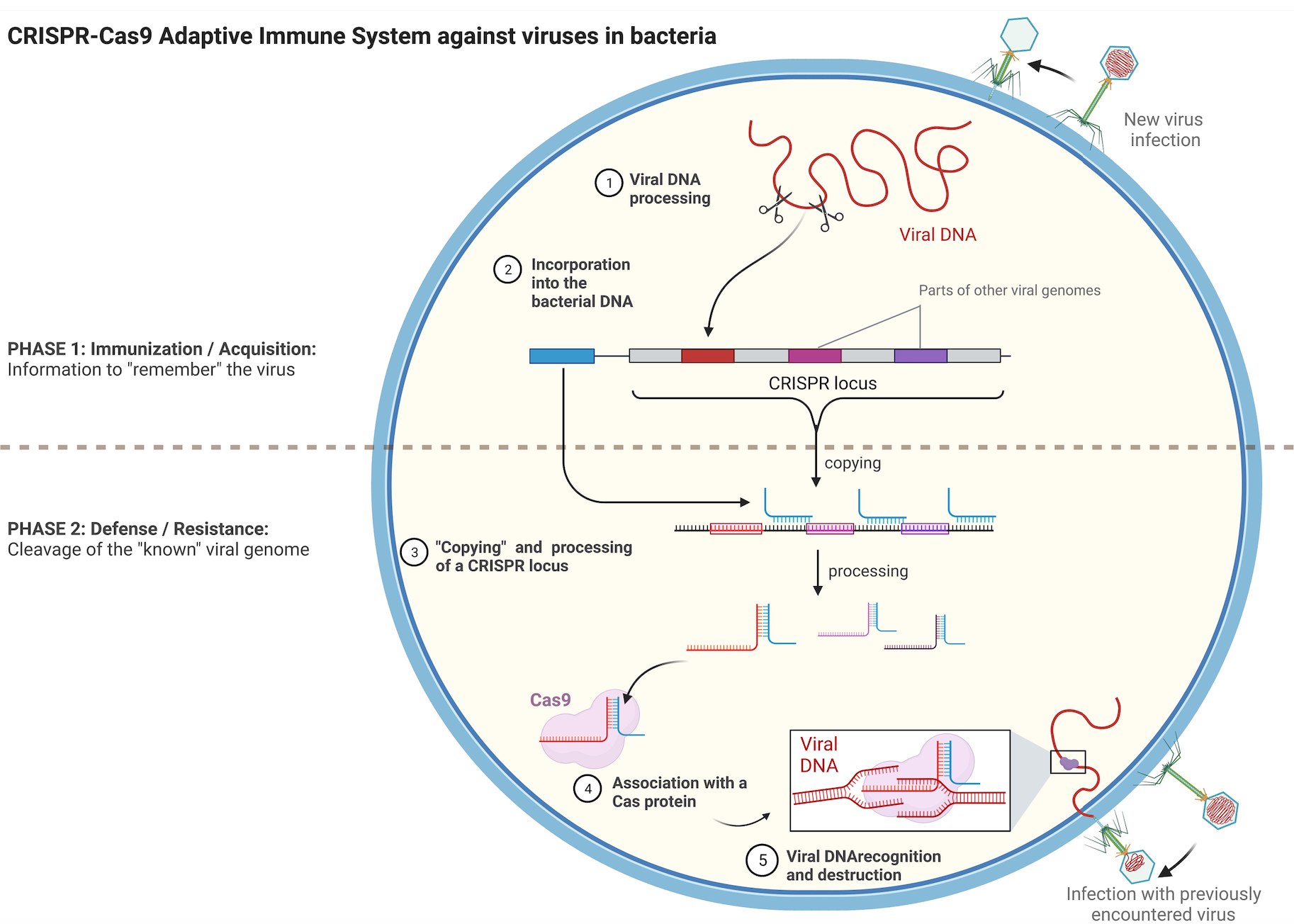
In bacteria, the CRISPR-Cas9 system is fundamental for protection against viral infections. Using a small RNA molecule called guide RNA, the CRISPR-Cas9 system acts as a nuclease, akin to DNA cutting scissors, destroying the viral genome. This system can be modified to edit genes in organisms beyond bacteria. The guide RNA is altered to direct the Cas DNA scissor to a gene of interest instead of a viral genome. This programmable gene-editing system has diverse applications, from basic biology research to disease treatments, and it earned the 2020 Nobel Prize in Chemistry for Emmanuelle Charpentier and Jennifer Doudna. Notably, scientists have recently traced the evolutionary origins of CRISPR-Cas9 to transposons.
In a recent study published in Nature, Columbia postdoc Chance Meers and his colleagues provided further insight into the evolution of the CRISPR-Cas9 nuclease system by examining how transposons use cellular nucleases to proliferate within genomes. They focused on a specific transposon system called insertion sequences (IS), which are bacterial transposons encoding only the genes necessary for transposon excision and movement to a new genomic location. This includes a transposase. However, many IS transposons contain an accessory gene that carries the information for a nuclease which functions similarly to the CRISPR-Cas system as an RNA-guided DNA cleaving enzyme.
The IS element does not need a nuclease for its movement, the transposase is sufficient to mobilize the transposon to a new location. So why are so many of the IS elements carrying those RNA-guided DNA scissors? While the mechanism by which IS elements are mobilized by their transposase is similar to a cut-and-paste process, leaving no copy at the original DNA location, Meers and colleagues discovered that the rate of excision is more efficient than the rate of integration. Without an additional system for transposon proliferation, the transposon would eventually be lost since not every excised copy manages to be reintegrated. The study of the authors revealed that the accessory CRISPR-like functioning nuclease guides a copy of the IS element back to its original location, generating two copies of the element—one at the original site and one at the newly inserted site. This changes the mechanism from cut and paste to cut and copy when the transposase function is complemented by the accessory nuclease, increasing the transposon copy number present in the genome, and thus serving for the transposon selfish proliferation (Figure 1).
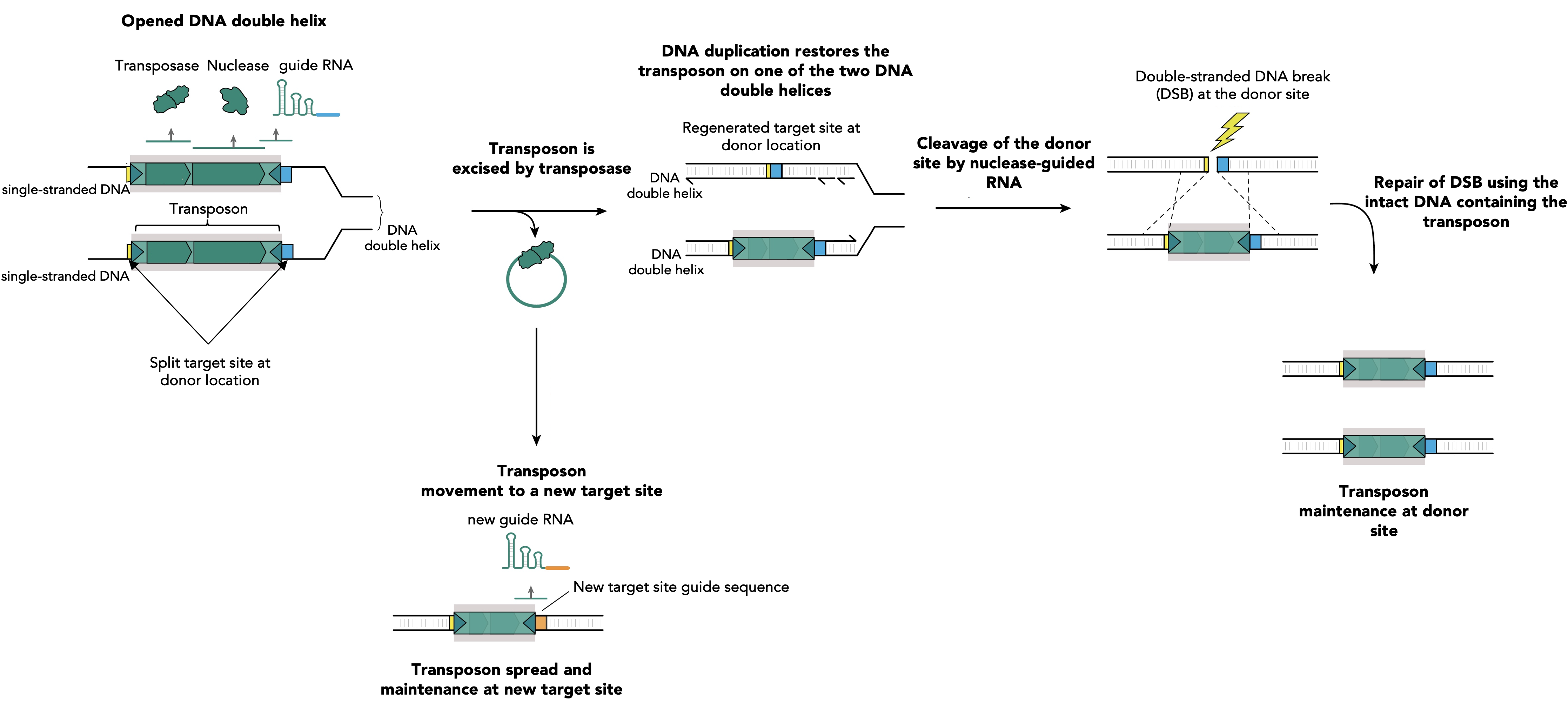 Figure 1. RNA-guided nucleases assist transposon survival via guiding specific breaks at donor sites and transposon restoring repair. During DNA replication the two strands of the DNA duplex are separated and each strand serves as a template for the synthesis of a new DNA molecule. Single stranded DNA facilitates the transposition mediated by the transposase of IS elements. The excision of the transposon leads to its loss at one of two DNA strands, while DNA replication restores the transposon on the other DNA strand. Simultaneously, transposon excision restores a target site at the transposon donor location which is specifically recognized by a guide RNA (blue segment). This guide RNA directs the accessory nuclease on the excision location and DNA cleavage occurs. The resulting double strand break (DSB) is lethal for the bacteria and its only chance of survival is to copy the information from the homologous newly synthesized intact DNA molecule that contains the transposon. This leads to reconstitution of the transposon at the donor site on the two newly synthesized DNA duplexes. Transposition at the new target sites will produce new guides, specific for the new insertion site location (orange segment), which will facilitate the future transposon spread and maintenance by identical mechanism.
Figure 1. RNA-guided nucleases assist transposon survival via guiding specific breaks at donor sites and transposon restoring repair. During DNA replication the two strands of the DNA duplex are separated and each strand serves as a template for the synthesis of a new DNA molecule. Single stranded DNA facilitates the transposition mediated by the transposase of IS elements. The excision of the transposon leads to its loss at one of two DNA strands, while DNA replication restores the transposon on the other DNA strand. Simultaneously, transposon excision restores a target site at the transposon donor location which is specifically recognized by a guide RNA (blue segment). This guide RNA directs the accessory nuclease on the excision location and DNA cleavage occurs. The resulting double strand break (DSB) is lethal for the bacteria and its only chance of survival is to copy the information from the homologous newly synthesized intact DNA molecule that contains the transposon. This leads to reconstitution of the transposon at the donor site on the two newly synthesized DNA duplexes. Transposition at the new target sites will produce new guides, specific for the new insertion site location (orange segment), which will facilitate the future transposon spread and maintenance by identical mechanism.
By developing powerful assays to track the movement of transposons within bacterial genomes, or other small DNA molecules called plasmids, as well as from one bacterium to another, Meers and his colleagues uncovered how those RNA-guided DNA cutting nucleases work. The authors’ discovery enhances our understanding of how proteins collaborate with RNA guides to target and edit genomes. In addition, the study unveils the original role of CRISPR’s nucleases from an evolutionary perspective before they were repurposed by the bacterial host genome to fight viruses, which was to serve the selfish propagation of transposons. Moreover, given the abundance of transposons in genomes, it is highly likely that other systems different from CRISPR-Cas9 and derived from transposon genes exist, waiting to be discovered and potentially harnessed, expanding the biotechnological tools available for programmable and specific genome engineering. Thousands of ancient transposons in bacterial genomes carry RNA-guided DNA nucleases that can potentially be programmed to cut DNA similarly to the CRISPR-Cas9 system.
Reviewed by: Trang Nguyen, Giulia Mezzadri