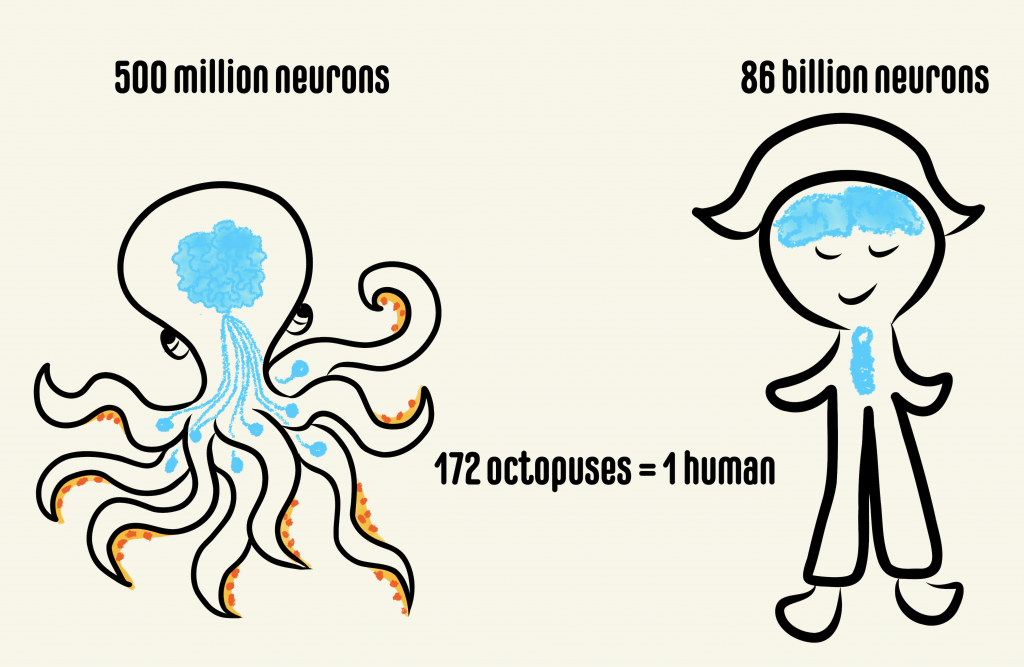
Anyone who watched the movie Arrival would not miss the conspicuous resemblance of the alien Heptapods to some of our own earthly beings – the octopuses. While serving as inspiration for alien creatures in movies, as clairvoyants in soccer World Cup or as savages in classic science fiction and mythology, cephalopods like octopuses and squids have been dubbed as one of the most intelligent creatures on the planet. There is a good reason for why cephalopods, particularly octopuses, have developed such a reputation. Octopuses have a striking organization of brain structure, different from that of any other studied organism. They have the largest nervous system among animals lacking a backbone, comprising a total of nine “brains”. Out of these, one is a major donut-shaped brain that contains ~200 million neurons surrounding the octopus’s food pipe, which is strangely located in the head! This brain communicates behavioral intricacies to the eight so-called mini-brains located within the arms, each containing ~40 million neurons. The central brain is responsible for executing complex behaviors like tool use, ability to plan for the future, shape-shift, camouflage, recognize individuals and solve complex puzzles. While the last common ancestor between octopuses and humans was about 680 million years ago, a recent surprising discovery showed that they both evolved to use the same molecules during development. Scientists discovered that genes that produced the camera-like eye in humans are the same ones that gave rise to the camera-like eye in octopuses. What’s more, these cephalopods have evolved complex brains that show behavioral innovation on par with a small primate.
Despite the potential that the octopus provides for understanding developmental biology, particularly the brain, the molecules that dictate how the mollusk’s brain is built are unknown. The common octopus, Octopus vulgaris, is specifically suited to address this question because it can produce thousands of small and transparent eggs in a single batch and scientists have recently mapped out most of its genes. Using O. vulgaris in studies led by Dr. Astrid Deryckere from Dr. Eve Seuntjens’s lab at KU Leuven in Belgium, the group set out to unravel these molecular mysteries. “If you would think of cephalopods as the primates of the sea, that have evolved a complex nervous system from a far more simple ancestral nervous system, surprisingly little is understood on the morphological and molecular mechanisms driving its development.”, said Dr. Deryckere. They approached the problem in two studies. In the first study, she established a system for controlled embryonic development, which enabled her to care for thousands of eggs without the need for the mother octopus. She used state of the art microscopy and recorded high-resolution images of octopus development from fertilization through hatching. This work from Dr. Deryckere and colleagues can now be used as an elaborate reference for cephalopod embryology.
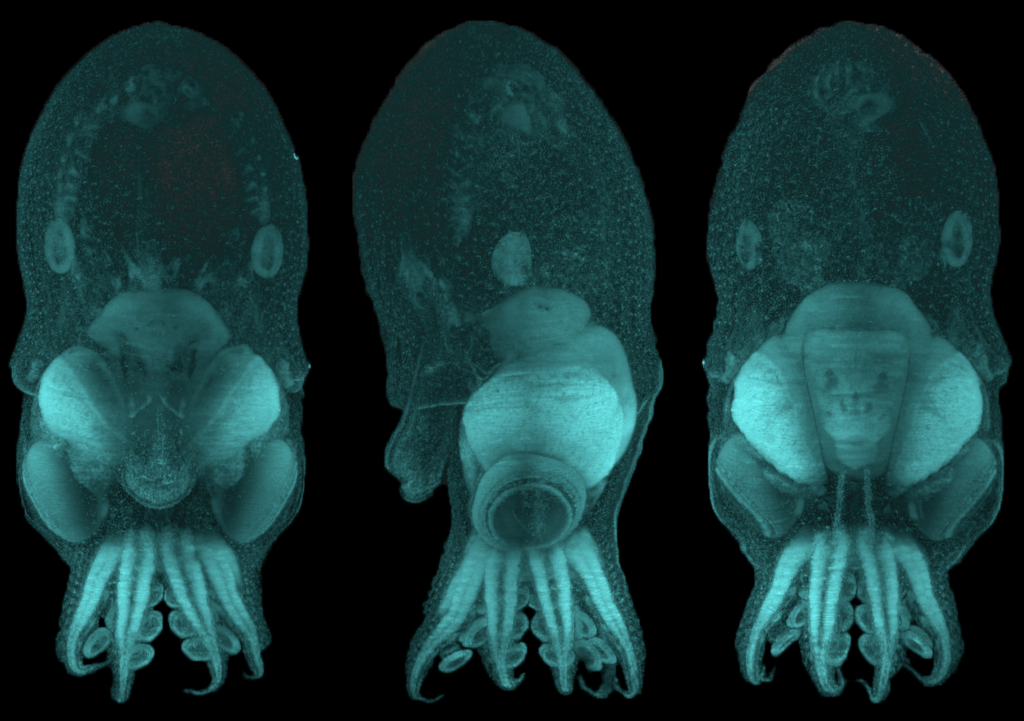
In the second study, Dr. Deryckere dug deep into the origins of the octopus brain. She used precise staging to track the precursor cells of neurons, or “neuronal progenitors”, that generate specialized neurons. Intriguingly, these progenitor cells appeared in structures called the lateral lips, that are unrelated to the brain and are located around the eyes. So, it appeared that neurons were first born in these structures and eventually migrated into the central brain – a possibility that prompted the authors to investigate further. They found that hundreds of thousands of neurons were created within octopus embryos even before hatching. To find out what genes are required for this unique way of making neurons, Dr. Deryckere used molecular markers and showed for the first time that newborn neurons travel long-distances to reach their final location in the brain. The results showed that the genes were expressed in the same order that vertebrates like humans do. By closely observing entire embryos in three dimensions during their growth, she found that neurons proceed through maturation while migrating from the lateral lips to an intermediate transition zone that finally leads to the brain.
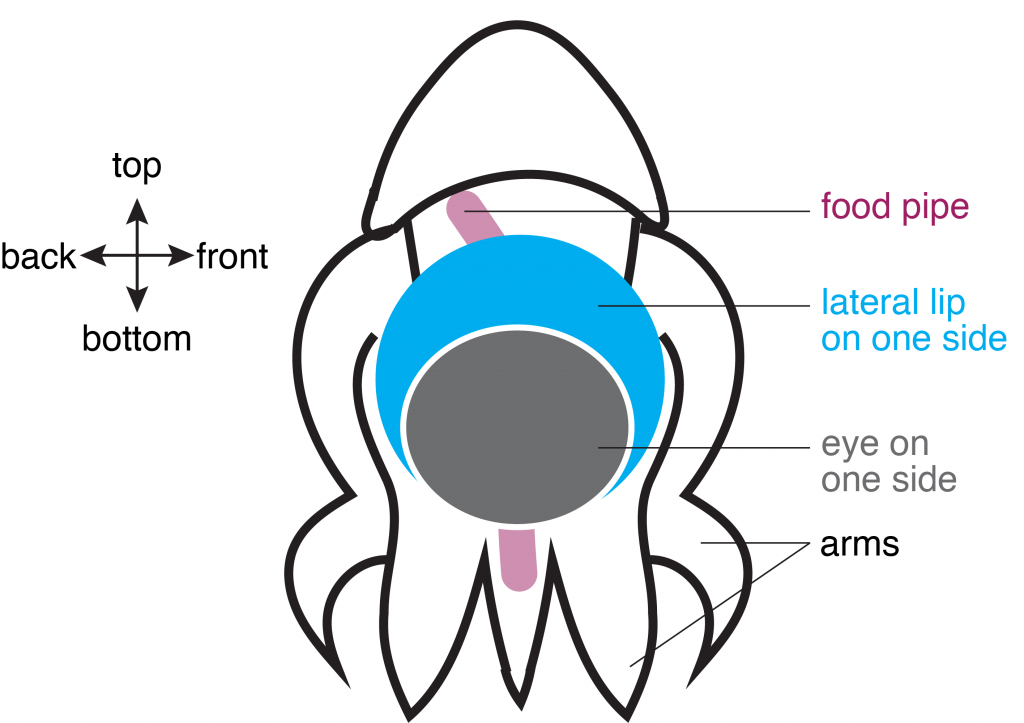
Using detailed molecular studies, the scientists now show support for the lateral lips harboring newly dividing neuronal cells in the embryo. This is unusual because unlike in human brains and many other organisms, the dividing cells are located outside the central brain. These dividing cells then unwaveringly make their way towards the final destination to maturity in the octopus central brain. “The migration is especially exceptional for invertebrates where neurons usually migrate only a few cell lengths”, noted Dr. Deryckere about the rarity of the observation.
This unique development in the octopus head and its interesting age-dependent arrangement of dividing cells and mature neurons only inspires further reverence for the cephalopod. While it continues to influence characters in pop culture, the glorious octopus and its brain hold even more promise in the real world. The octopus brain’s cognitive ability has galvanized a new age of artificial intelligence, leading to the construction of flexible robotics and prosthetics, but at the same time, is pushing scientists and philosophers to tackle the important question of how an intelligent life form is defined.
Dr. Astrid Deryckere is currently a postdoc in the lab of Maria Tosches in the Department of Biological Sciences at Columbia University. Her focus remains on brain development but she has transitioned to working on an animal with a backbone – the salamander.