Alphaviruses can infect both vertebrate and invertebrate animals. Their transmission between species and individuals occurs mainly via mosquitoes. These viruses are small, spherical, and have a genome composed of a single strand ribonucleic acid (RNA) in the “positive-sense”. The alphaviral life cycles and their RNA genome amplification (replication) have been studied since their discovery in 1953. However, the very initial events of viral genome replication have remained unknown.
Positive-strand RNA viruses genome can be directly translated into viral proteins with the participation of factors and structures provided by the invaded host cell. However, in order to amplify the viral genome and to produce new viral particles during the virus propagation, the positive RNA strand has to be converted to its complementary negative strand by an enzyme that is encoded in the viral genome. This enzyme uses RNA as a template to synthesize RNA, a so-called RNA-dependent RNA polymerase (RdRp). RdRp are used during replication of the genome to synthesize a negative-sense antigenome that is then used as the template to create a new positive-sense viral genome, necessary for the future viral progeny and viral propagation (Figure 1).
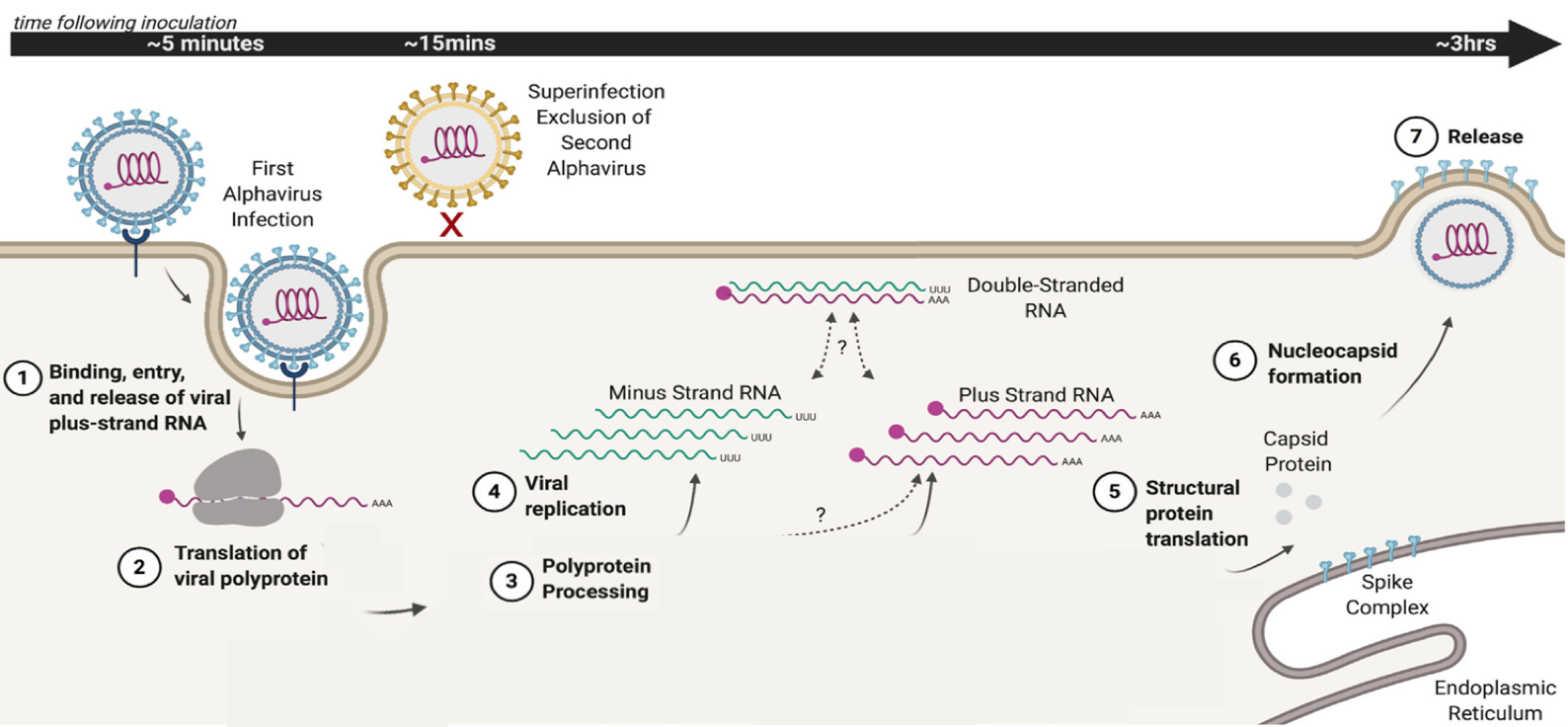
Figure 1. Overview of the alphavirus life cycle. Alphaviruses enter the cell by recognizing a cell receptor, followed by release into the host cell of the viral plus-strand genome (1). The genome serves as a template carrying the information for production of a fused version of viral proteins (viral polyprotein, 2). This polyprotein is cleaved to different combinations (not shown) constituting an RNA-dependent RNA polymerase, and two forms of protein complexes required for viral replication (3 and 4). The consecutive cleavage of the polyprotein has been shown to influence transitions in production between the full-length minus-strand RNA, the genomic plus strand, as well as of another form of viral RNA (not shown) required for subsequent viral particles (nucleocapsid) assembly and release (5 and 6). Figure adapted from the original paper.
A phenomenon known as superinfection exclusion has been previously observed, where infection by one virus can block the infection of a subsequent homologous virus. This form of viral competition protects the virus to complete its reproduction without the need to share the cell’s resources with homologous viruses or with its own progeny. One of the mechanisms of superinfection inclusion can be by reducing the host cell receptors that the virus uses to recognise and enter into the cell. However, such changes in the cells are thought to take place several hours upon infection and for some viruses the phenomenon of superinfection exclusion has been observed as soon as just 15 minutes of the first infection (Figure 1). This rapid competitive behavior was observed over 40 years ago. This mechanism providing such rapid protection over a secondary infection is very beneficial, especially considering the ability of the virus to enter cells within minutes. However, its causes, as well as the very earliest stages of alphaviral replication and whether the two processes are linked has remained unclear.
Previous studies of the alphavirus’s life cycle have mainly used populations of infected cells. The use of recently developed single cell-based methods allows to overcome several limitations of population-level studies. For example, the classic population-based studies have shown the average growth of the virus over time across millions of cells and have revealed that the first release of viral progeny can be detected as early as 3–4 hours post infection (Figure 1). However, there is an inherent cell-to-cell variability in the infection spreading in a group of cells. Use of single-cell analyses in biology has shown how the variability of individual cells can be masked by the overall population’s behavior and how variability between individual cells contribute to viral growth and spreading kinetics. An important challenge on how the dynamics of early replication could affect the competitive interactions is the lack of sensitivity on low-abundance targets during early infection. In order to capture the dynamics of the earliest stages of replication, it is necessary to utilize an approach with sufficient sensitivity to simultaneously measure individual molecules of multiple viral RNA species at low abundance.
The recent work published from Columbia postdoc Zakary Singer and his colleagues presents a new quantitative detailed characterization of the initial replication activity of members of the alphavirus genus, Sindbis virus. The study consists in analyzing the viral genome biology at the level of individually affected cells and not in a group of cellular population. The authors used quantitative live single cell imaging technique to follow and measure the viral replication in real time upon infection as well as to elucidate how these contribute to the rapid exclusion of a superinfecting alphavirus. Singer and colleagues observed that the rapid onset of viral RNA synthesis as a passive superinfection exclusion mechanism could contribute to this advantage. Furthermore, a mathematical model of exponential viral growth in a resource-limited environment appeared consistent with the measurements of viral replication. The authors also investigated whether there is a bidirectional inhibition between two viruses in the same cell, by experimental measurements and a mathematical modeling of competitive growth using parameters estimated from single-virus infection experiments. The results from both methods suggested that the superinfecting virus is equally able to reduce replication levels of the first virus and that the cell appears to have fixed carrying capacity that sets up the combined replication level of the two viruses. Due to the speed of Sindbis replication would strongly disadvantage the second virus and reduce the second virus’s replication, showing the importance of intrinsic growth kinetics in alphaviral superinfection exclusion.
The work by Singer and colleagues also allowed to shed light on classic questions remaining in alpha virology and suggested a revised model of early replication wherein both plus- and minus strands are made at a similar rate during early infection in contrast to previous claims that initially the positive strand RNA production is predominant. Additionally, the paper provides one of the earliest detections of alphaviral replication, as well as a new framework for understanding early replication and the resulting exclusionary phenomenon. Finally, the work hints on how in the future the complex interplay with innate immunity and stochasticity will be broadly relevant to the study of many infectious diseases, and how quantitative models might lead to improved antivirals. Check out more from the original publication.
Edited by: Sam Rossano and Trang Nguyen