Macrophages, a type of immune cells, are an integral part of our body’s defense system. The term macrophage comes from two Greek words – makro meaning big and phagien meaning eat, which makes them the “big eaters”. And boy, do they love to eat! Some things that they like to chomp on include bacteria and other foreign substances, dying and dead cells, and cancer cells, thus, acting as the body’s cleanup system. This process of eating is not only important for defending against foreign pathogens but is also essential for cleaning up cell debris and maintaining normal bodily functions.
Macrophages typically encapsulate their food by surrounding it with cell extensions, then engulf it and digest it. Check out some cool videos of macrophages eating some bacteria here. This process of eating is typically called “phagocytosis”. However, the term for macrophages eating dying cells is called “efferocytosis”. This term is derived from the Latin word efferre which translates to “take to the grave” or “to bury”. When this mechanism of disposal of cellular corpses goes wrong, the rotting dead cells can lead to inflammation that damages the surrounding tissue. This can lead to many diseases, including coronary artery disease, chronic obstructive pulmonary disease, cystic fibrosis, and rheumatoid arthritis. In a recent publication from the Tabas lab, Dr. Kasikara and Columbia postdoc Dr. Schilperoort explore the molecular mechanisms that underlie impaired efferocytosis and how that leads to the formation of dangerous plaques in the arteries that supply blood to your heart. The buildup of these plaques leads to a condition called coronary artery disease which remains the leading cause of deaths in the United States, causing about 1 in 4 deaths.
Significant advances in genomic sequencing in the past few years have led to the discovery of several mutations that are often correlated with the occurrence of coronary artery disease in patients. One of these mutations is in a gene encoding a protein called PHACTR1. However, because the mutation is present in a part of the gene outside of where the protein-coding sequence lies, it was unclear if this mutation disrupted efferocytosis by disrupting the function of PHACTR1. PHACTR1 regulates the ability of various cell types to expand, contract, and move. While the ability of macrophages to execute these motions is required to engulf or eat cells, whether PHACTR1 is involved in this process in macrophages and thereby macrophage efferocytosis was not known. In this study, the authors made two important discoveries. Firstly, they found that PHACTR1 is essential for macrophage efferocytosis. Secondly, they found that the mutation decreases the expression levels of PHACTR1. The authors investigated more and established that PHACTR1 is important for maintaining an activated version of a motor protein called myosin which is required for cellular movement. Thus, lower levels of PHACTR1 hamper the ability of macrophages to eat dead cells by disrupting cellular movement. This contributes to the buildup of dying cells in our arteries and a consequent increase in the risk of heart attack and stroke.
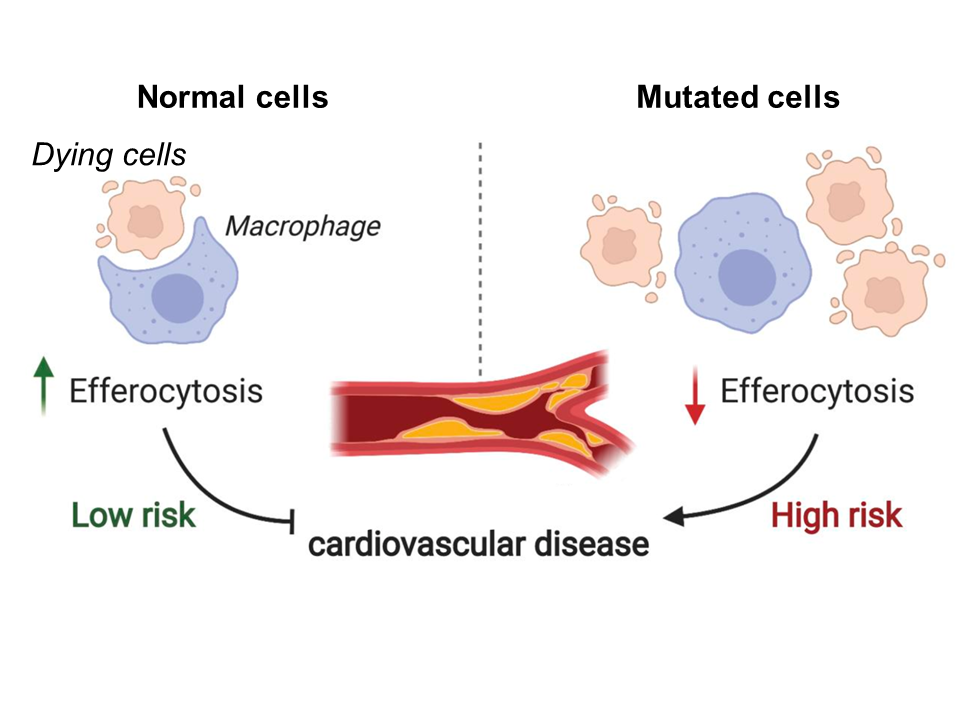 Fig 1. Model depicting the relationship between efferocytosis and risk of coronary artery disease. Reduced levels of efferocytosis lead to insufficient clearance of dead cells and consequent plaque formation in the arteries. Figure adapted from Kasikara, JCI 2021.
Fig 1. Model depicting the relationship between efferocytosis and risk of coronary artery disease. Reduced levels of efferocytosis lead to insufficient clearance of dead cells and consequent plaque formation in the arteries. Figure adapted from Kasikara, JCI 2021.
The results from this study provide novel insights into the role of PHACTR1, myosin, and other associated proteins in the pathogenesis and progression of coronary artery disease. Before this study was performed, we only knew that there was a correlation between an increased risk of heart disease and a mutation in PHACTR1 gene. The authors performed rigorous experiments and demonstrated that the mutation changes PHACTR1 production and that this causes heart disease. This information is extremely valuable as it provides a basis for designing future therapies. For example, increasing PHACTR1 production artificially may be an effective strategy for treating coronary artery disease. As defective macrophage efferocytosis is also involved in the pathogenesis of many other diseases, this study has direct implications for the discovery of new treatment paradigms for these diseases as well.