Acute lymphoblastic leukemia (ALL) is an aggressive form of cancer arising from malignant transformation of immature cells that were otherwise fated to become white blood cells, or lymphocytes. ALL occasionally affects adults but is more commonly a pediatric cancer: children under the age of 5 have the highest risk of being affected. Upon diagnosis, it is possible to treat ALL with an aggressive regimen of multiple chemotherapy drugs that is successful for over 80% of patients. Unfortunately, when tumors reappear after initial treatment, in relapse, the cases become extremely difficult to treat. Additionally, the cellular landscape of ALL in relapse has a high degree of genetic heterogeneity and variability between different patients. Oncologists and cancer biologists suspect it is this genetic complexity that makes ALL in relapse especially hard to fight in the clinic.
In a study published in Nature Cancer last year, Dr. Jessie Brown and colleagues set out to improve outcomes for patients with ALL by clarifying how mutational complexity in relapsed tumors interacts with chemotherapy drugs to resist treatment. To this end, the team first employed extensive genetic sequencing on ALL samples collected upon diagnosis, remission, and relapse in order to identify the mutational landscape that distinguishes relapse tumors from the others. Samples were collected from 175 patients in total – 149 from pediatric cases and 26 from adults. Next, in order to functionally characterize the mutations they identified, the authors used a genome-wide genetic screening strategy to identify drug–gene interactions and determine why the relapse-specific mutational landscape is less responsive to chemotherapy. The screen was carried out in a representative model ALL cell line using CRISPR, a genetic editing tool used to specifically activate or inhibit the expression of single genes.
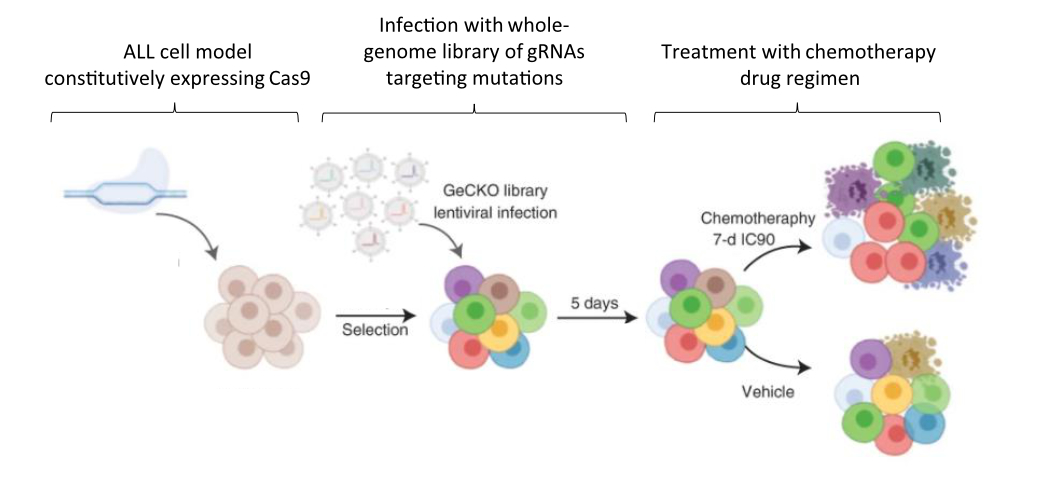
Between these two approaches, the team succeeded in characterizing relapse-specific mutations that arise during the administration of chemotherapy itself, a process known as clonal evolution. Additionally, the number of mutations they identified increased with patient age at diagnosis, a finding that allowed the researchers to establish that the most recent commonality between the mutant cells present at diagnosis versus later at relapse often develops early, years before the leukemia is officially diagnosed. Importantly, this finding is consistent with the hypothesized fetal origin of many pediatric ALLs, which postulates that chromosomal abnormalities leading to cancer are already present at birth. It is also consistent with the higher rate of relapse previously observed in adult patients.
When the mutations uniquely acquired during relapse were further investigated using CRISPR screening in an ALL model cell line, a strong positive selection was revealed for those that conferred chemotherapy resistance. By using CRISPR to manipulate the expression of genes affected by each mutation of interest and assessing how the ALL model cells fared in each experiment, the researchers were able to analyze the relationships between the effect of the mutation and application of each drug. Of the drugs investigated, functional overlaps in the cellular mechanisms mediating the activity of each were identified between several groups of them. The significance of this finding is two-fold. First, it helps researchers and medical providers understand why the presently used multi-drug regimen might be effective for ALL in the first place. Second, it suggests that other drugs acting via similar mechanisms of action could be effective treatments in the future. Moving forward, ALL in relapse might be treated not just with combinatorial chemotherapy, but with specific combinations, doses, and schedules of drugs that meet the personalized genetic vulnerabilities of specific ALL cases.
One inhibitor tested in the study’s cell-based CRISPR screen, an inhibitor called ABT-199, also known as Venetoclax, is already being tested for inclusion as a new therapeutic. If approved, it could become part of the arsenal of drugs used to compose personalized chemotherapeutic cocktails for patients with ALL in relapse. According to co-first author Dr. Brown, “it is currently in Phase I/II clinical trials for relapsed ALL and other malignancies and we hope that this work and our follow-up studies can further underscore the mechanisms of action of this inhibitor in combination with commonly used chemotherapies.”
Altogether, this study identified a number of mutations that make relapsed ALL distinct from ALL before treatment with chemotherapy and functionally characterized the interactions of these mutations with multiple chemotherapy drugs. While ALL is not a common cancer – it accounts for less than 0.05% of all incidences of cancer in the United States – those affected must be treated with aggressive chemotherapy that can negatively affect the lives and health of patients in many ways. Because of this, understanding how to better target ALL at diagnosis and treatment-resistant ALL in relapse is a high priority for researchers. The findings of this work help to identify targets for reversing chemotherapy resistance and improving treatment outcomes for pediatric and adult patients alike.
Dr. Jessie Brown is a postdoctoral research fellow in the Ferrando Lab at Columbia University Irving Medical Center studying therapeutic resistance in relapsed acute lymphoblastic leukemia.